Associate Professor of Chemistry & Biochemistry Emeritus
452 Coulter Hall
662-915-5339 | mmossing@olemiss.edu
EDUCATIONAL AND PROFESSIONAL BACKGROUND
B.S, Michigan State University, 1980
Ph.D., University of Wisconsin, 1985
Postdoctoral Fellow, Massachusetts Institute of Technology, 1986-1990
RESEARCH INTERESTS
Biophysical chemistry; DNA binding protein structure and function; combinatorial mutagenesis; protein engineering and expression; circular dichroism, fluorescence, and NMR spectroscopy; thermodynamics and kinetics of protein folding and macromolecular recognition; the coupling of protein folding and assembly to DNA recognition
RESEARCH SUMMARY
Proteins bind to specific DNA sites and to each other to control gene expression. These regulatory complexes direct the construction and maintenance of cells and organisms from their genomic blueprints. Genetics and molecular biology give us the tools to identify transcriptional regulatory proteins and their DNA binding sites and map out the control circuits that drive living systems. Structural biology enables the examination of these proteins and their complexes with DNA in atomic detail. Work in my laboratory is aimed at characterizing the thermodynamic and kinetic properties of these protein-DNA interactions. The conditions under which we can make biochemical measurements on purified components in the laboratory are often far removed from the crowded, complex microcosm of a living cell. In order to verify our extrapolations we make measurements under various solution conditions and on proteins and DNA sites that have been engineered or manipulated to test specific hypotheses. The ultimate test of our success will be in developing mathematical models that describe the behavior not only of natural regulatory systems but systems built from modified components that have been characterized in vitro.
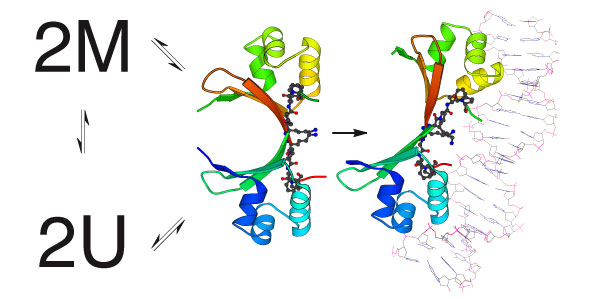 Cro folding and dimer assembly are coupled to DNA recognition The Cro repressor of bacteriophage lambda is one of the simplest and best characterized of any DNA binding protein. It helps to control a simple binary genetic switch that toggles between two lifestyles for bacteriophage lambda and the life or death of lambda’s bacterial host. We have shown that a major limitation in Cro’s ability to bind to DNA is in the formation of the protein dimers that are necessary for DNA binding. Studies of the kinetics and thermodynamics of protein folding and dimer assembly are ongoing. Engineered variants of Cro that are trapped either in the dimeric state or the monomeric state or that fold at different rates and assemble to different extents will will help in completing our understanding of this simple genetic circuit.
Cro folding and dimer assembly are coupled to DNA recognition The Cro repressor of bacteriophage lambda is one of the simplest and best characterized of any DNA binding protein. It helps to control a simple binary genetic switch that toggles between two lifestyles for bacteriophage lambda and the life or death of lambda’s bacterial host. We have shown that a major limitation in Cro’s ability to bind to DNA is in the formation of the protein dimers that are necessary for DNA binding. Studies of the kinetics and thermodynamics of protein folding and dimer assembly are ongoing. Engineered variants of Cro that are trapped either in the dimeric state or the monomeric state or that fold at different rates and assemble to different extents will will help in completing our understanding of this simple genetic circuit.
RECENT PUBLICATIONS
Maity H, Mossing MC, Eftink MR. Equilibrium unfolding of dimeric and engineered monomeric forms of lambda Cro (F58W) repressor and the effect of added salts: evidence for the formation of folded monomer induced by sodium perchlorate. Arch Biochem Biophys. Feb 1;434(1):93-107.
Jia H, Satumba WJ, Bidwell GL 3rd, Mossing MC. Slow assembly and disassembly of lambda Cro repressor dimers. J Mol Biol. Jul 29;350(5):919-29.
Mollah AK, Stennis RL, Mossing MC. Stability of monomeric Cro variants: Isoenergetic transformation of a type I’ to a type II’ beta-hairpin by single amino acid replacements. Protein Sci. 2003 May;12(5):1126-30.
4: Satumba WJ, Mossing MC. Folding and assembly of lambda Cro repressor dimers are kinetically limited by proline isomerization. Biochemistry. Dec 3;41(48):14216-24.
